[version 13][version 12][version 11][version 10][version 9][version 8][version 7][version 6]
6.3 Sampling and monitoring data
There are a set of general post-processing functions for sampling data across the domain for graphs and visualisation. Several functions also provide data in a single file, in the form of time versus values, that can be plotted onto graphs. This time-value data can be monitored during a simulation with the foamMonitor script.
6.3.1 Probing data
The functions for probing data are boundaryCloud, internalCloud and probes as listed in section 6.2.1.9. All functions work on the basis that the user provides some point locations and a list of fields, and the function writes out values of the fields are those locations. The differences between the cloud and probe functionality are as follows.
- probes identifies the nearest cells to the probe locations and writes out the cell values; data is written into a single file in time-value format, suitable for plotting a graph.
- boundaryCloud and internalCloud interpolate field data to the probe locations, with the locations being snapped onto boundaries for boundaryCloud; data sets are written to separate files at scheduled write times (like fields). data.
Generally probes is more suitable for monitoring values at smaller numbers of locations, whereas the cloud functions are typically for sampling at large numbers of locations.
As an example, the user could use the pitzDaily case set up in section 6.2.3. The probes function is best configured by copying the file to the local system directory using foamGet.
foamGet probes
13#includeEtc "caseDicts/postProcessing/probes/probes.cfg"
14
15fields (p U);
16probeLocations
17(
18 (0.01 0 0)
19);
20
21// ************************************************************************* //
The configuration is completed by adding the #includeFunc directive to functions in the the controlDict file.
functions
{
#includeFunc probes
... other function objects here ...
}
6.3.2 Sampling for graphs
The singleGraph function samples data for graph plotting. To use it, the singleGraph file should be copied into the system directory to be configured. We will configure it here using the pitzDaily case as before. The file is simply copied using foamGet.
foamGet singleGraph
14start (0.01 0.025 0);
15end (0.01 -0.025 0);
16fields (U p);
17
18// Sampling and I/O settings
19#includeEtc "caseDicts/postProcessing/graphs/sampleDict.cfg"
20
21// Override settings here, e.g.
22// setConfig { type midPoint; }
23
24// Must be last entry
25#includeEtc "caseDicts/postProcessing/graphs/graph.cfg"
26
27// ************************************************************************* //
The configuration is completed by adding the #includeFunc directive to functions in the the controlDict file.
functions
{
#includeFunc singleGraph
... other function objects here ...
}
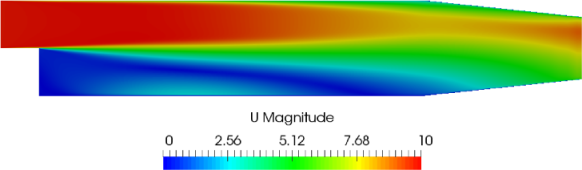
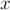 -component of velocity,
-component of velocity, 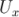 in the last time 296, by running gnuplot and
plotting values.
in the last time 296, by running gnuplot and
plotting values. gnuplot
gnuplot> set style data linespoints
gnuplot> plot "postProcessing/singleGraph/296/line_U.xy" u 2:1
The formatting of the graph is specified in configuration files in $FOAM_ETC/caseDicts/postProcessing/graphs. The sampleDict.cfg file in that directory contains a setConfig sub-dictionary as shown below.
9interpolationScheme cellPoint;
10
11setFormat raw;
12
13setConfig
14{
15 type lineUniform; // lineCell, lineCellFace
16 axis distance; // x, y, z, xyz
17 nPoints 100;
18}
19
20// ************************************************************************* //
This shows that the sampling is at uniform distribution of 100 points and that the
axis labelling is based on the distance from the start point. Instead the user may
wish to sample at the mid-point of each cell, using the 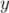 -ordinate for the axis
labelling. This can be done by override the settings in setConfig in their
singleGraph file, as shown below.
-ordinate for the axis
labelling. This can be done by override the settings in setConfig in their
singleGraph file, as shown below.
14start (0.01 -0.025 0);
15end (0.01 0.025 0);
16fields (U p);
17
18// Sampling and I/O settings
19#includeEtc "caseDicts/postProcessing/graphs/sampleDict.cfg"
20
21// Override settings here, e.g.
22setConfig
23{
24 type lineCell;
25 axis y;
26}
27
28// Must be last entry
29#includeEtc "caseDicts/postProcessing/graphs/graph.cfg"
30
31// ************************************************************************* //
Running simpleFoam produces the graph in Figure 6.6.
6.3.3 Sampling for visualisation
The surfaces and streamlines functions can be used to generate files for visualisation. The use of streamlines is already configured in the pitzDaily case. The surfaces function is configured by copying the surfaces file to the system directory using foamGet.
foamGet surfaces
 -direction.
-direction.
17#includeEtc "caseDicts/postProcessing/visualization/surfaces.cfg"
18
19fields (p U);
20
21surfaces
22(
23 zNormal
24 {
25 $cuttingPlane;
26 pointAndNormalDict
27 {
28 normalVector $z;
29 }
30 }
31);
32
33// ************************************************************************* //
The function can be included as normal from the by adding the #includeFunc directive to functions in the the controlDict file. Alternatively, the user could test running the function using the solver post-processing by the following command.
simpleFoam -postProcess -func surfaces
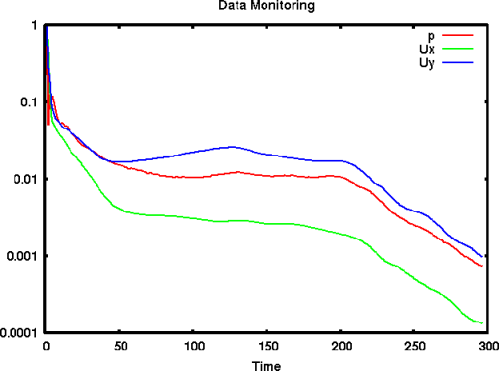
6.3.4 Live monitoring of data
Functions like probes produce a singe file of time-value data, suitable for graph plotting. When the function is executed during a simulation, the user may wish to monitor the data live on screen. The foamMonitor script enables this; to discover its functionality, the user run it with the -help option. The help option includes an example of monitoring residuals that we can demonstrate in this section.
Firstly, include the residuals function in the controlDict file.
functions
{
#includeFunc residuals
... other function objects here ...
}
foamInfo residuals
foamGet residuals
It is advisable to delete the postProcessing directory to avoid duplicate files for each function. The user can delete the directory, then run simpleFoam in the background.
rm -rf postProcessing
simpleFoam > log &
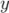 -axis on the residuals file as follows. If the command is executed before
the simulation is complete, they can see the graph being updated live.
-axis on the residuals file as follows. If the command is executed before
the simulation is complete, they can see the graph being updated live.
foamMonitor -l postProcessing/residuals/0/residuals.dat

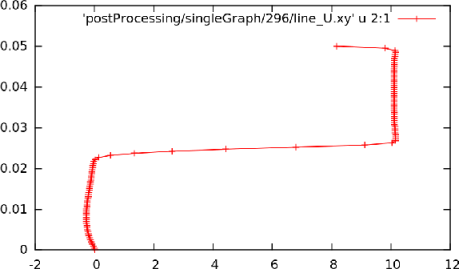
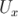 at
at 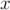 = 0.01, uniform sampling
= 0.01, uniform sampling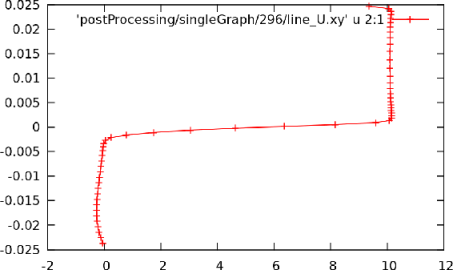
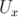 at
at 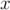 = 0.01, mid-point sampling
= 0.01, mid-point sampling